top of page
The HSF professional system was specifically designed to handle NGS data both from small, medium or large gene panels including WES/WGS. To do so, it precomputed the impact of mutations on splicing signals and stored results in the HSF database.
The user can access mutations either one-by-one or by a handful through the website of by directly submitting a VCF file through an API. The result is provided as a JSON format in order to be implemented in any local pipeline. Depending on the server load and the mutation type, the speed can reach 10,000 mutations/s.

Access to HSF predictions using the website. Various interactive tables allow the access to predictions.
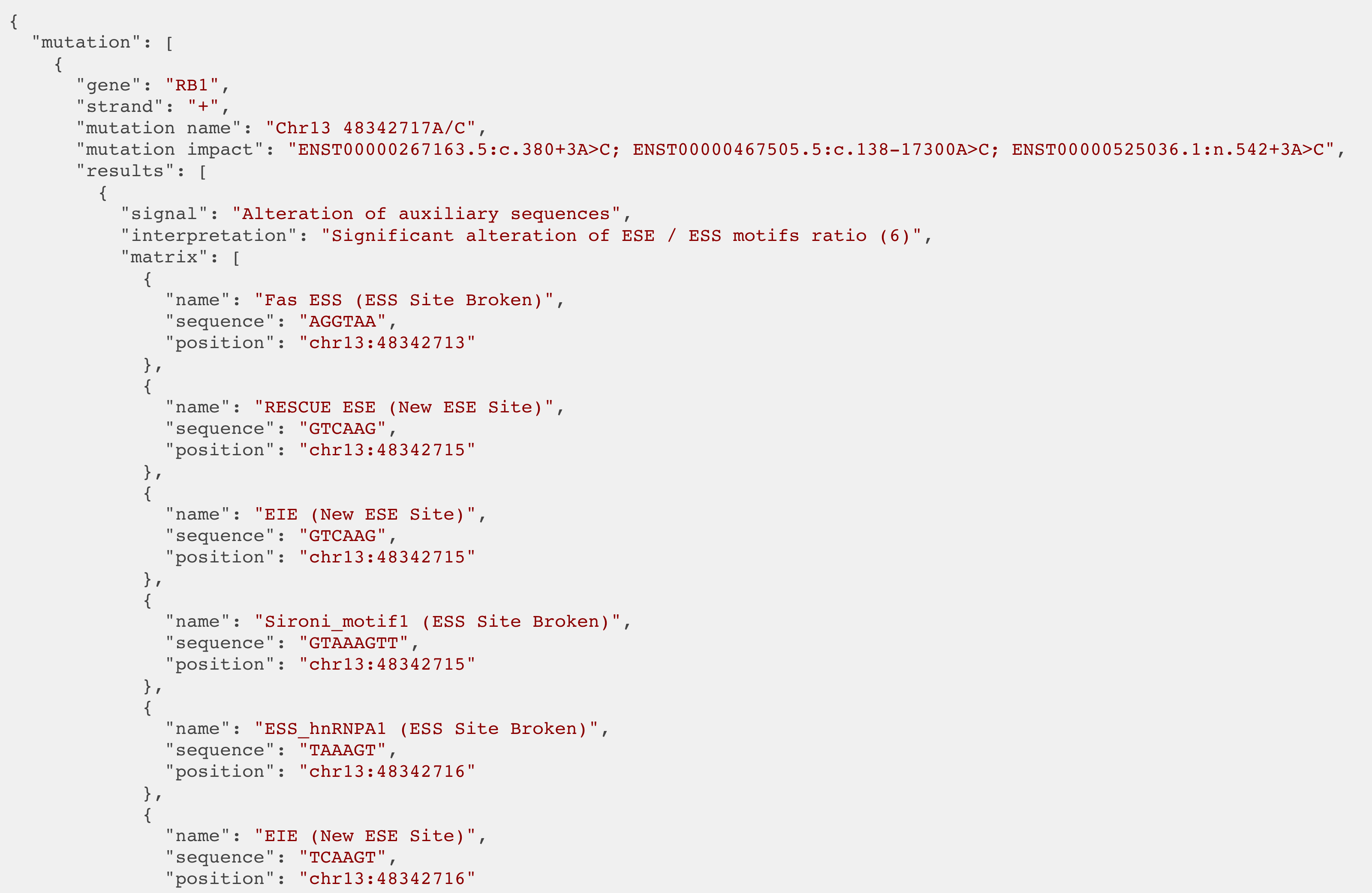
HSF predictions returned to the API using a JSON format..
bottom of page
